surface - an R package for fitting macroevolutionary adaptive landscapes
Travis' miscellaneous R scripts: I use R for most of my statistical analysis and simulation models, and have gathered a few scripts that might be useful to others. I can't guarantee they're bug-proof or interpretable, but I've tried to provide enough annotation that they’ll make sense. Send me an email if you find errors or have questions about anything.
The Ψ (psi) model of speciational trait evolution.
This code fits the Ψ model, which estimates the rate of 'speciational' evolution concentrated at the nodes of a phylogenetic tree relative to the rate of gradual evolution (Ingram 2011, Proc. R. Soc. B). This model has been updated and implemented as a part of the motmot.2.0 R package (github page). This tutorial shows the steps for using the code to fit the Ψ model and to use it to test hypotheses about whether clades or traits differ in the extent of speciational vs. gradual evolution.
The old code is here: psi_function.R along with the original example tree and depth habitat data for 66 species of rockfish (Sebastes). Note that this code contains a minor error in how Ψ is calculated, which is corrected in the updated implementation.
Script for simulating the eco-FOB model of coextinctions in food webs (Ingram & Steel 2010, J. Theor. Biol.). Also, a .csv file describing the Chesapeake Bay food web that we used as an example.
Script used to carry out null model analyses of the distribution of traits and phylogenetic relatedness in communities (Ingram & Shurin 2009, Ecology). To run the analyses on our rockfish data set, I've also included the script file and four data files (trait data, tree, community composition and community descriptions) in a .zip archive.
Function for calculating the diet variation indices WIC, BIC, TNW and WIC/TNW (Roughgarden 1972, Am Nat; Bolnick et al. 2002, Ecology) for continuous prey data (e.g. prey sizes) when numbers of prey items vary among individuals (Ingram et al. 2011, PLoS ONE). Some of this code was incorporated into the RInSp R package.
Software and Code
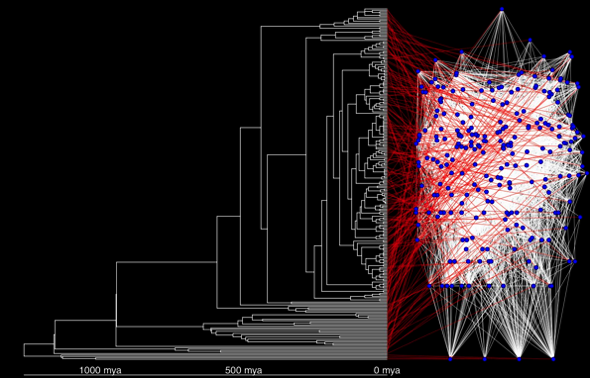
Fun with R graphics: Phylogenetic history and trophic links of a large reef food web
