Thursday 25 August 2022 9:48am
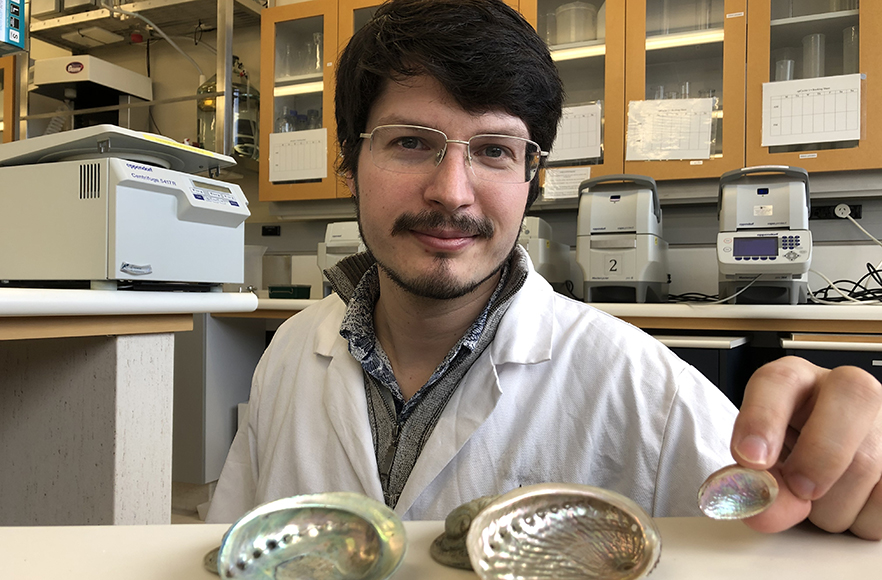
PhD candidate Kerry Walton holds shells from each of the three paua/abalone species from mainland New Zealand. The smallest of the shells, an adult Haliotis virginea (virgin paua), was the focus species of his study. Photo: University of Otago.
A new DNA extraction method has made hundreds of thousands of shells in museum collections available for study.
The international group of researchers behind the technique hopes it will lead to improved conservation approaches for endangered and never-before-seen-alive molluscs.
Lead author Kerry Walton, PhD candidate in the University of Otago’s Department of Zoology, says there are about 200,000 different species of mollusc, making them the second most diverse animal group after arthropods.
The shells of clams, oysters, mussels, and snails are robust and can last in some environments for hundreds or even thousands of years.
“About half of living mollusc species have never been seen or collected alive and are only known from their shells.
“While there are hundreds of thousands of mollusc specimens in museum collections in Aotearoa New Zealand, only a few thousand, a tiny fraction, are suitable for conventional genetic research approaches.
“We managed to develop a combination of methods that improved the retrieval of DNA and sequencing of genomes from mollusc shell by up to 350-fold over previous methods, and we did so in a way that proved reliable and relatively affordable, without destroying the specimens,” he says.
The researchers used the new technique in a study, published in Molecular Ecology Resources, to analyse New Zealand abalone shells. The results provided better understanding around how different regional species relate to one another.
The technique enables novel questions to be answered, for the good of scientific understanding and conservation.
“Enabling researchers to use historically collected shells rather than relying on freshly collected specimens would significantly increase the number of specimens and species available for genetic research.
“This would enable, for example, conservation genetic approaches to be applied to endangered or never-before-seen-alive molluscs. A better understanding of invertebrates is critical to understanding soil or water health, and ecosystems as a whole.”
He believes natural history collections, often housed in museums, are extremely important but often understated.
“We can now look at how genetic diversity in molluscs have changed through time, from before human arrival in Aotearoa to the present day and use that information to better understand evolutionary processes and provide evidence-based conservation management guidelines for our taonga species.”
Publication details:
Application of palaeogenetic techniques to historic mollusc shells reveals phylogeographic structure in a New Zealand abalone
Kerry Walton, Lachie Scarsbrook, Kieren J. Mitchell, Alexander J. F. Verry, Bruce A. Marshall, Nicolas J. Rawlence & Hamish G. Spencer
Molecular Ecology Resources
https://onlinelibrary.wiley.com/doi/10.1111/1755-0998.13696
The Conversation:
The researchers have written a piece for The Conversation. You are welcome to republish this, with appropriate attribution.
Empty mollusc shells hold the story of evolution, even for extinct species. Now we can decode it.
For more information, please contact:
Kerry Walton
Otago Palaeogenetics Laboratory
Department of Zoology
University of Otago
Ellie Rowley
Communications Adviser
University of Otago
Mob +64 21 278 8200
Email ellie.rowley@otago.ac.nz
