Programme directors
Associate Professor Brian Monk, Programme leader
Associate Professor Geoffrey Tompkins, Deputy programme leader
Programme staff
Key research staff, postgraduate students and collaborations
Preventing oral diseases caused by microorganisms
Oral microbes cause disease in a large proportion of our population. In the Molecular Microbiology programme we study how the diseases are caused and how we can prevent or overcome them. We use biochemical, molecular biological, and microbiological techniques within the Molecular Biosciences Laboratory to investigate oral and other microbial diseases.
Molecular Biosciences Laboratory
Interested in joining our programme?
How do bacteria overcome host defences?
We're investigating how periodontal bacteria acquire the haem they require for growth, as preventing this access may help prevent periodontal disease. We're also looking at how bacteria colonise and invade dentinal tubules, which can lead to endodontic infections. We work on finding models of how to effectively combat these infections.
Unique oral microbiomes
A team is using next-generation DNA sequencing technology to analyse the bacterial diversity associated with oral health and disease (severe dental caries and periodontal disease). Next-generation DNA sequencing is also being used to sequence the genomes of two oral bacterial species:
An antimicrobial-producing strain of Streptococcus salivarius
A strain of a new species, Streptococcus trichosurus, isolated from the oral cavity of the New Zealand-adapted brushtail possum Trichosurus vulpecula.
Drug resistance in oral fungi
Oral fungi can cause mucosal and systemic infections. Our research team has discovered major mechanisms of azole and echinocandin resistance in oral fungi. We are currently screening for drugs to overcome azole resistance. The drug screening platform is also being applied to other, novel, antifungal drug targets and to drug targets implicated in other human diseases and parasitic nematodes.
Improved antifungal drugs
Oral fungi can develop resistance to the widely used azole antifungal drugs. We have recently crystallized and resolved the structure of the azole drug target Erg11p (also known as Cyp51) from baker's yeast Saccharomyces cerevisiae. We are using this structure to guide the design of improved antifungal drugs.
Drug resistance in tumour cells
Human ATP-binding cassette (ABC) transporters are responsible for the drug resistance of some tumour cells. Our team is improving the expression of human ABC transporters in yeast strains so they can be used for functional analysis and in screening for drug discovery.
More information
Our Molecular Microbiology research programme profile (linked at right) contains more details on our current research projects, collaborations funding highlights and recent publications.
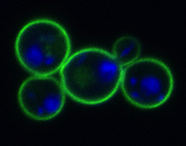
Fluorescence microscopy image of Saccharomyces cerevisiae yeast cells with plasma membrane protein Pdr5p tagged with green fluorescent protein and the nucleus stained with DAPI (blue).

Fluorescence microscopy image of Candida albicans hyphae and yeast cell walls stained with calcafluor white.
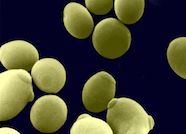
Scanning electron microscopy image of Candida albicans yeast cells.
More information
Molecular microbiology programme profile
(Extract from our 2019-2020 SJWRI Research Report)
Key research staff, postgraduate students and collaborations
